Don Teng
Scientific software developer

Baltic3
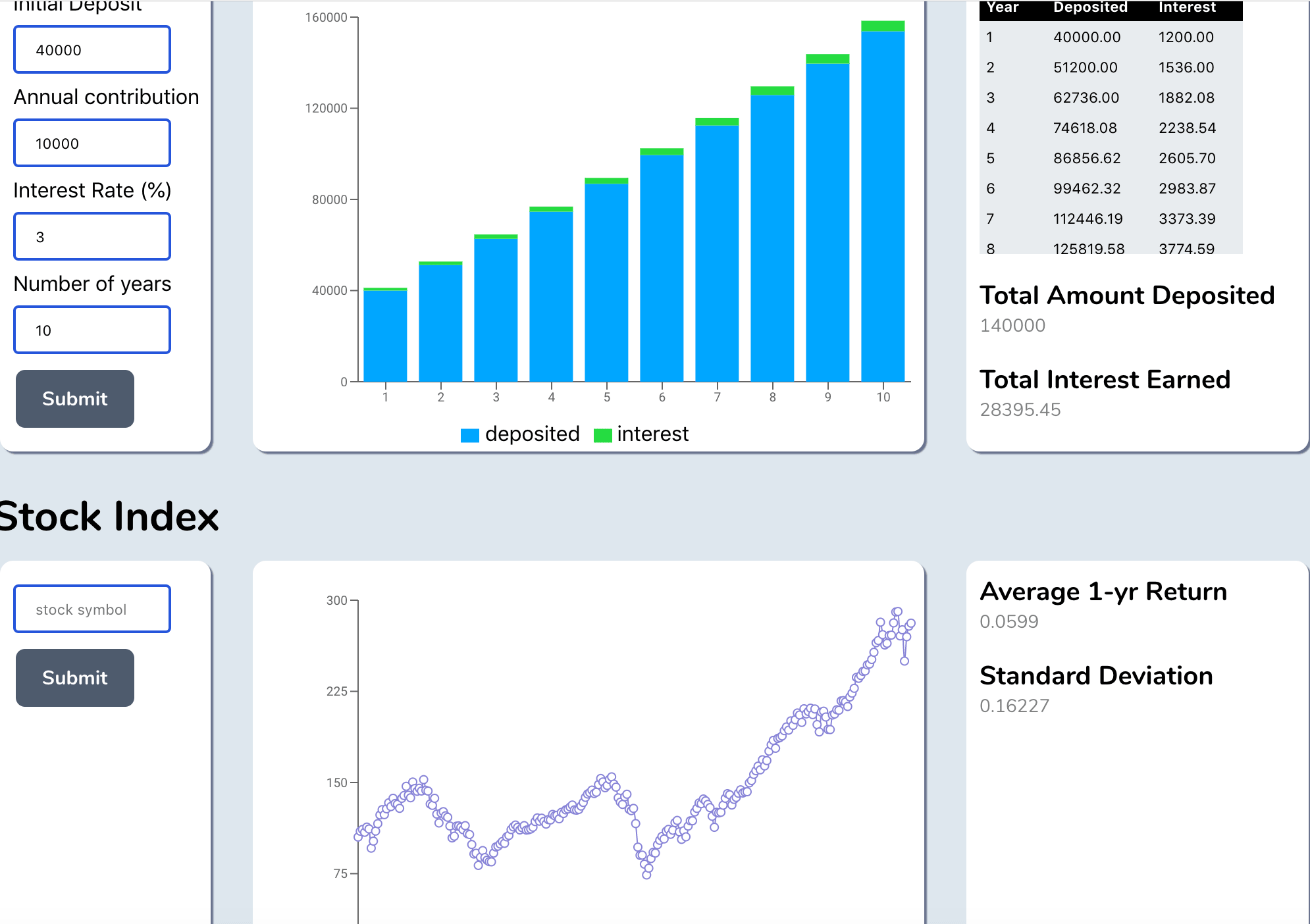
Finance Portfolio Dashboard
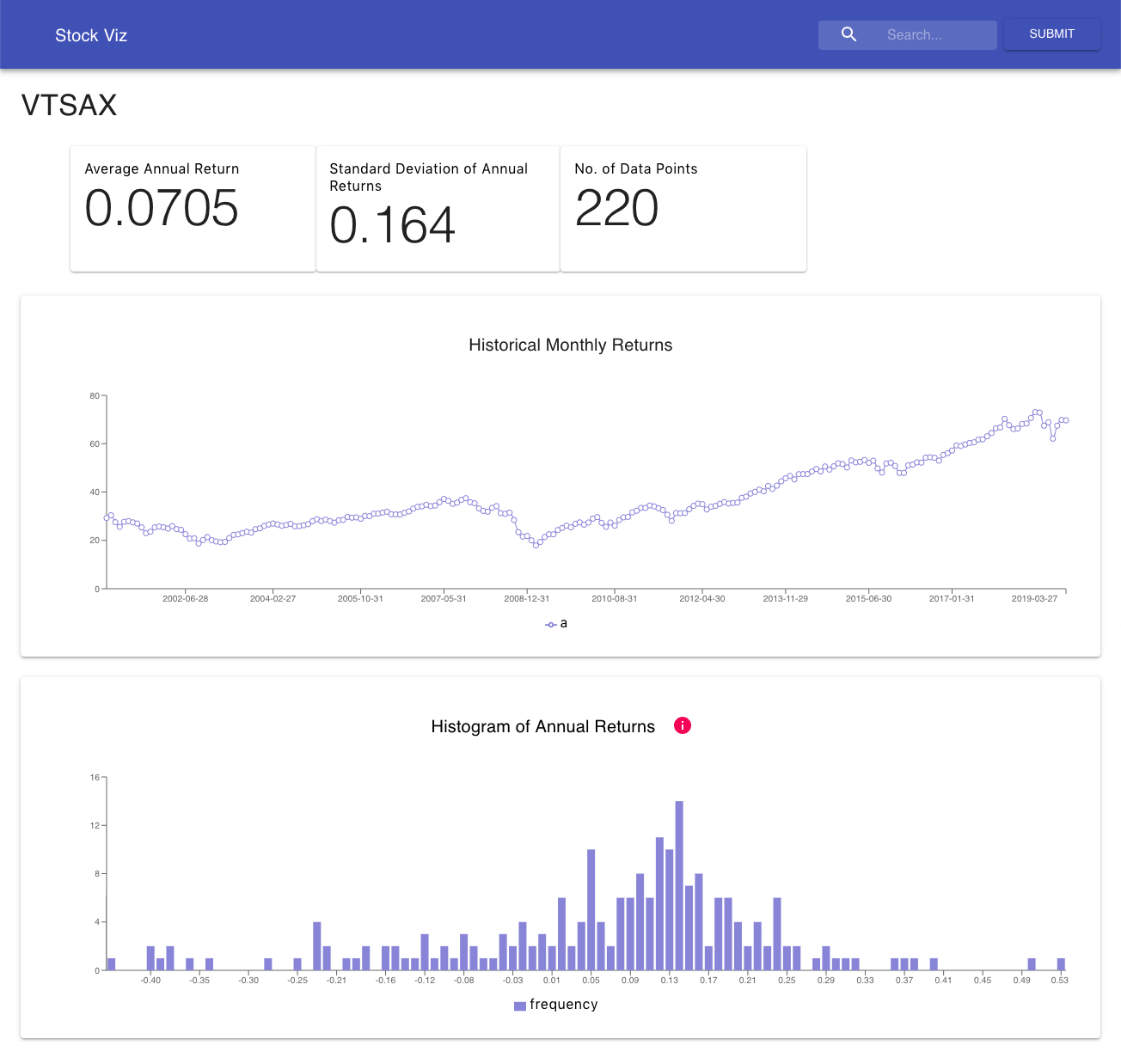
Stock Visualization Dashboard

Hello, REST API World
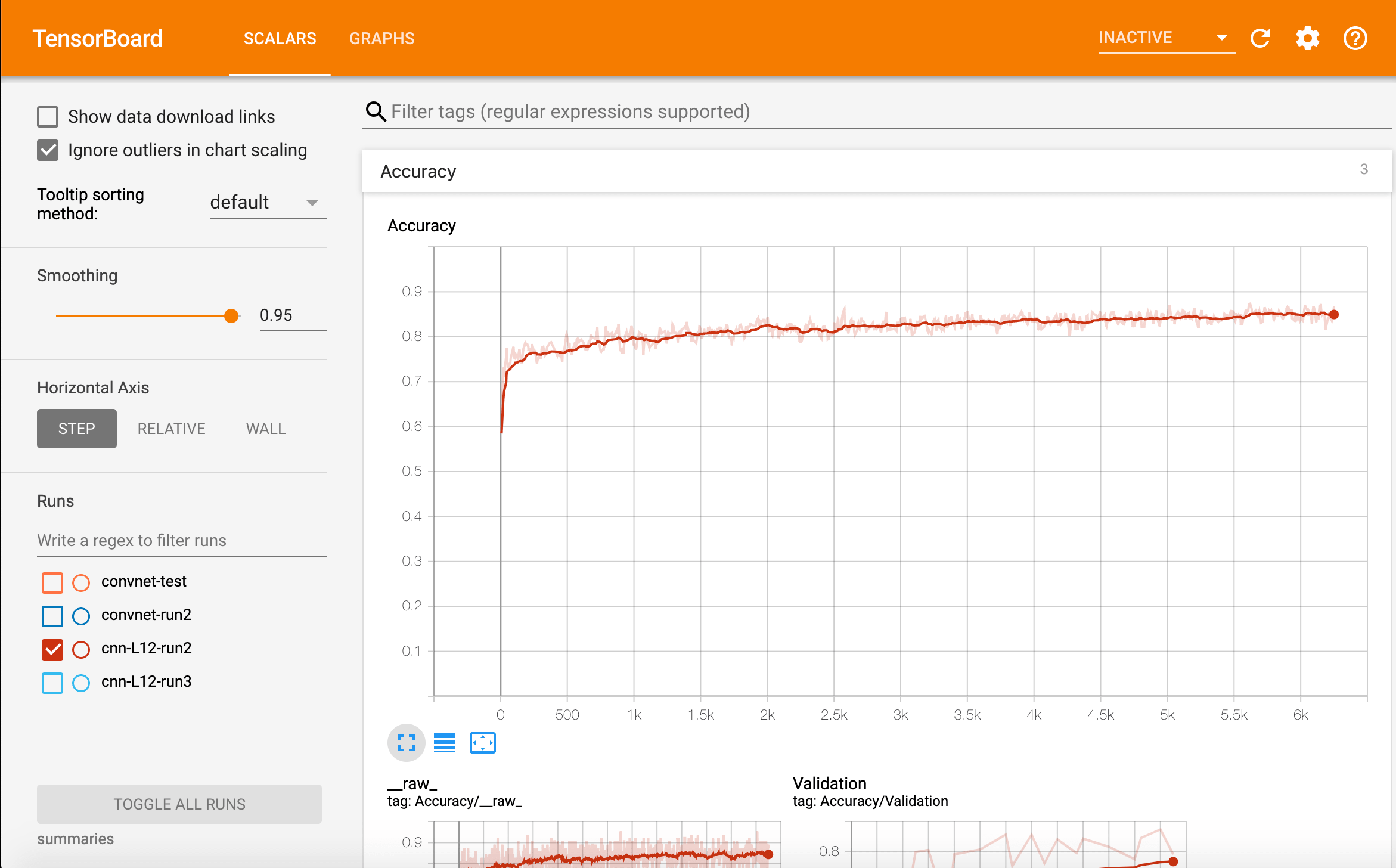
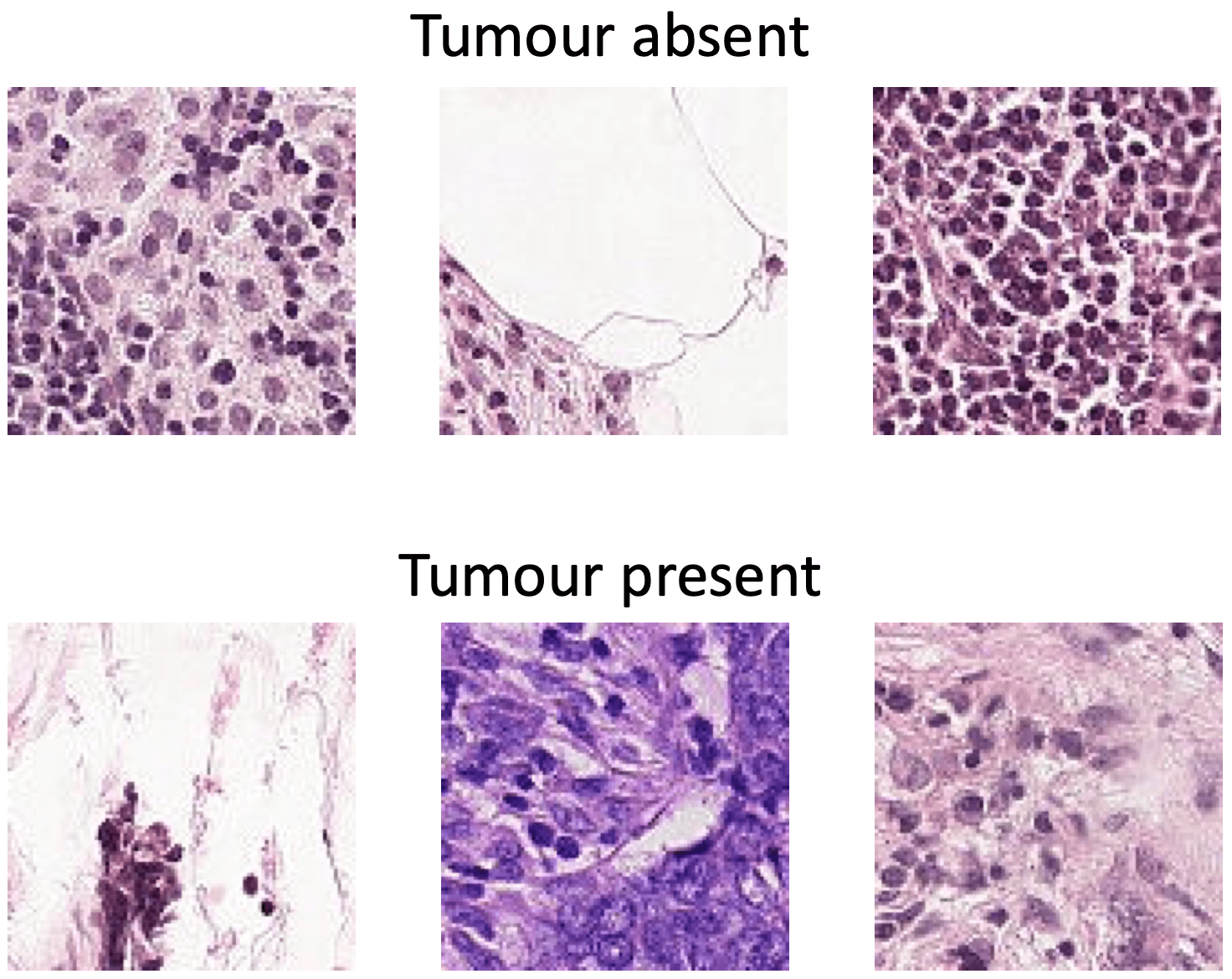
Identifying Metastatic Cancer from Small Image Scans